


File -> ReOpen (Ctrl+Shift+O) [reopen-file]
Reload the already loaded bio-network from the original file. If the current buffer is modified, a confirmation box is first popped.
File -> Insert (Ctrl+I) [insert-file]
Add to the right of the currently edited bio-network, the bio-network stored in a SBML, Metatool or a native format file. The file selection box is popped to get the name of the file to add.
File -> Insert Picture [insert-image]
Insert a picture (image file: jpg or png) in the currently selected arc or node label at the current cursor position.
File -> Save (Ctrl+S) [save-file]
Save the currently edited bio-network to the original file in SBML format if the file extension is 'xml' or in native format if the extension is 'net'. The current buffer is marked as non-modified.
File -> Save as (Ctrl+Shift+S) [save-as]
Save the currently edited bio-network into a file in SBML format if the file extension is .xml, or in native format if the extension is .net. The file selection box is popped to get the name of the file. The current buffer is then marked as non-modified.
File -> Export to HSIM [export-to-hsim]
Translate the currently edited bio-network to a configuration file suitable for the HSIM simulation system.
File -> Export to Metatool [export-to-metatool]
Translate the currently edited bio-network to a configuration file suitable for the METATOOL system.
File -> Export to PDF [export-to-pdf]
Export the currently edited bio-network to a PDF file. The file selection box is popped to get the name of the file.
File -> Save settings [save-settings]
Save the current values of the settings in the ~/.netdraw_rc file. These include:
File -> Save User Manual [save-user-manual]
Save the Silicell Maker user manual in a HTML file. The system's file selection dialog box is popped to choose the file name.
File -> Portrait/Landscape [print-orientation]
Set the main orientation for the printer output. Defaults is portrait (vertical).
File -> Print (Ctrl+P) [print]
Print the currently edited bio-network. The system's print dialog box is popped to select the printing settings.
File -> Quit (Ctrl+Q) [quit]
Quit Protocell Maker. If the current bio-network is modified but not saved, the confirmation box is popped.
Edit -> Undo (Ctrl+Z) [undo-command]
Undo the previous commands that modified the currently edited bio-network. Everything is undoable, even loading a new file!
Edit -> Redo (Ctrl+Y) [redo-commandd]
Redo the previously undone command.
Edit -> Find Reactant (Ctrl+F) [search-reactant]
Incremental search the reactants by name. The top and bottom arrows navigate between the occurrences found.
Edit -> Select all (Ctrl+A) [select-all-nodes]
Select all the nodes of the bio-network.
Edit -> Cut (Ctrl+X) [cut-selection]
Cut (and copy to the clipboard) the currently selected bio-network items.
Edit -> Copy (Ctrl+C) [copy-selection]
Copy to the clipboard the currently selected bio-network items.
Edit -> Paste (Ctrl+V) [paste]
Paste to the right of the currently edited bio-network the subnetwork stored in the clipboard. If there is only text in the clipboard, it can be pasted only into a label.
Edit -> Duplicate (Ctrl+Shift+V) [duplicate-selection]
Duplicate the currently selected bio-network items. The duplicate subnetwork become selected and so can be moved using a mouse drag. In the example below, the reaction p3 -> m1 has been duplicated and is selected.
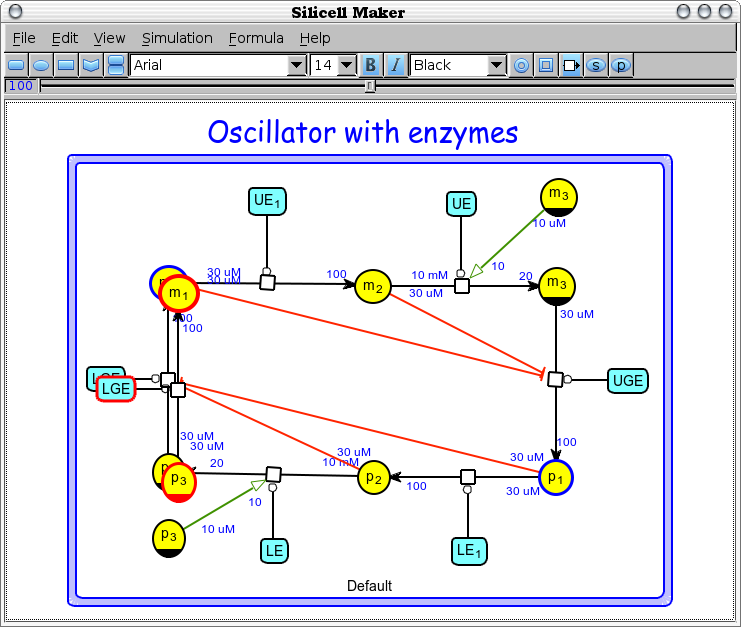
Edit -> Align nodes (Ctrl+J) [align-nodes]
Move the nodes (reactants and enzymes) of the currently edited bio-network in order to get the approximately horizontal and/or vertical reaction arcs perfectly horizontal and/or vertical.
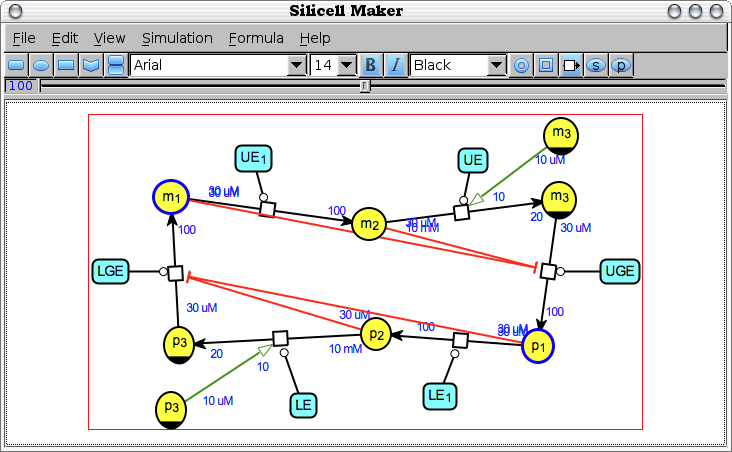
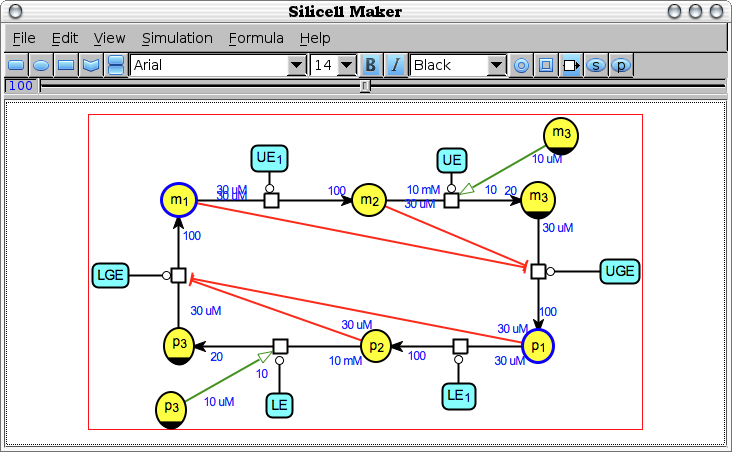
Edit -> Share clones [share-clones]
Find, in the currently edited bio-network, the nodes that represent the same biochemical species and change their type and graphical shape (according to the SBNG standards) to explicitely represent the same species. Conversely, the species appearing only once get their type and shape reset to normal.
Edit -> Color reactants [color-reactants]
Change the background colour of the substrate / product metabolites according to their status: input, internal, or output.
Edit -> Fog selection [fog-selection]
Display in low light intensity the subpart of the current bio-network that correspond to the current selection. All the reactions and reactants that can be reached from the any part of the selected subnetwork are fogged, unless they are involved in some other reactions.
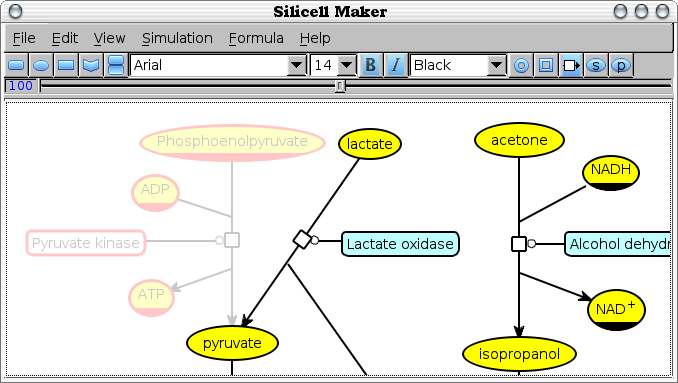
In the example above, the Phosphenol pyruvate is selected and all the components of the reaction it is part of, and the reaction itself are also blurred. Note that the pyruvate is not blurred since it is involved in other reactions.
Edit -> Unfog selection [unfog-selection]
Display in normal light intensity the subpart of the current bio-network that correspond to the current selection. All the reactions and reactants that can be reached from the any part of the selected subnetwork are un-fogged.
View -> Zoom in (Ctrl++) [zoom-in]
Enlarge by 5% steps (max 200%), the display of the current bio-network.
View -> Zoom out (Ctrl+-) [zoom-out]
Reduce by 5% steps (min 10%), the display of the current bio-network.
View -> Zoom reset (Ctrl+0) [reset-zoom]
Reset the display size of the current bio-network (to 100%).
View -> Show boundary [show-boundary]
Show/Hide a red rectangle corresponding to the bounding box of the current bio-network.
View -> Show mouse help [show-mouse-help]
Show/Hide the online contextual help about the mouse commands:

The 3 mouse buttons are shown as columns. The first line indicates what happens when there is no modifier, when Shift is used and when Ctrl is used. The line corresponding to the actual modifier used is enlighted and show what would happen if a mouse button would be pressed.
This display changes according to what is the object currently under the mouse pointer.
View -> Show kinetics [show-kinetics]
Show/Hide the reactions kinetics of the current bio-network.
View -> Show stoichiometry [show-stoichiometry]
Show/Hide the reactions stoichiometries of the current bio-network.
View -> Show grid [show-grid]
Show/Hide a grid that helps the user to align the nodes of the current bio-network. When moving a node, its center snaps to the grid edges.
View -> Smart move [smart-move]
Toggle the smart moving of the main reactants of a reaction (Mouse-1 Drag).
View -> Smart dialogbox [smart-dialog-box]
Switch between the standard popup dialog boxes and a faster keyboard-only version that uses a textfield inserted at the top of bio-network window. When used for a file selection a ls style window is displayed to help the file selection.
Formula -> Reactions/gate [netgate-set-depth]
Set the maximal number of reactions of the logic gates that NetGate will look for.
Formula -> Extract Gates [netgate-extract-gates]
Launch NetGate to build the database of enzymatic logic gates that will be used to translate boolean formulas to enzymatic networks. The file selection box is popped to get the name of the SBML file that contains the metabolic network from which the gates will be extracted.
Formula -> Enter Formula [netbuild-bool-to-network-translate]
Pop the formula dialog box to specify the boolean function to be implemented by an enzymatic network. This dialog box proposes the lists of all the possible input and output metabolites. Five pushbuttons can be used to enter one of the 3 basic boolean operators (AND, OR, NOT) and the opening and closing parenthesis to group boolean clauses. The formula can also be directely entered in a text input field.
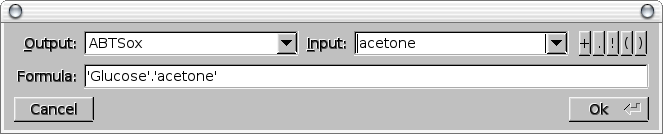
Then NetBuild is launched to assemble the enzymatic networks that implement the boolean formula using the previously built logic gates database. The first implementation found is loaded in the main window.
Formula -> Previous Implementation [netbuild-next-implementation]
Load the enzymatic networks of the previous implementation of the boolean formula found by NetBuild.
Formula -> Next Implementation [netbuild-previous-implementation]
Load the enzymatic networks of the next implementation of the boolean formula found by NetBuild.
Help -> About Silicell Maker [about-Silicell Maker]
Show the version and credits window.
Help -> Documentation [pop-help-box]
Show this documentation window.

Pointer in a molecule Mouse-1 Click [select-reactant]
Clear the current selection and select the pointed molecule.
Pointer on a reaction Mouse-1 Click [select-reaction]
Clear the current selection and select the pointed reaction.
Pointer on a catalyse arc Mouse-1 Click [select-reaction]
Clear the current selection and select the pointed catalyse arc.
Pointer in a text field Mouse-1 Click [edit-text]
Put the editing cursor on the text line at the pointed character and enter the line edit mode.
Pointer in the background Mouse-1 Click [clear-selection]
Deselect the currently selected molecules / reaction arcs / compartments.
Pointer in a molecule Ctrl+Mouse-1 Click [add-to-selection]
Add the pointed molecule to the current selection.
Pointer on a reaction Ctrl+Mouse-1 Click [add-to-selection]
Add the pointed reaction to the current selection.
Pointer on a catalyse arc Ctrl+Mouse-1 Click [add-to-selection]
Add the pointed catalyse arc to the current selection.
Pointer on a reaction Shift+Mouse-1 Click [add-sub/prod]
Add the (only one) molecule currently selected as a new substrate of the pointed reaction. The type-change context menu (Mouse-3 Click when the pointer is on the substrate arc) can be then used to make this molecule a product of the reaction.
Pointer on a reaction Shift+Mouse-1 Click [show-stoichiometry]
When the selection is empty, the stoichiometry labels of the reaction are created. They can be changed at any time.
Pointer on a reaction Shift+Mouse-3 Click [show-kinetics]
When the selection is empty, the kinetics fields of the reaction are created. They can be changed at any time.
Pointer in a molecule Shift+Mouse-1 Click [add-reaction]
Create a new reacton with the (only one) molecule currently selected as substrate and the pointed molecule as product. The type-change context menu (Mouse-3 Click when the pointer is on the reaction arc) can be then used to change type of the reaction.
Pointer in a molecule Shift+Mouse-1 Click [change-main-prod.]
Deconnect the main product of the currently selected reaction and connect it to the pointed molecule.
Pointer in a molecule Shift+Mouse-1 Click [swap-main-reactant]
Swap the main reactant (substrate or product) of the currently selected reaction with the pointed reactant
Pointer in a molecule Mouse-3 Click [change-species]
Pop a menu to change the type of the pointed molecule.
Pointer in the background Mouse-1 Drag [rubber-band]
Draw a rubber band selection rectangle where all the nodes located inside become selected. When the mouse button is released, the system enters a new state where the next mouse-1 drag will move those selected nodes.
Pointer in a selected molecule Mouse-1 Drag [move-selection]
Move the currently selected molecules. The arcs follow the molecule's moves
Pointer in the background Mouse-3 Drag [scroll-network]
Scroll the bio-network inside the viewing window.
Pointer in a kinetics/stoichiometry reaction field Mouse-3 Drag [move-label]
Move manually the kinetic (or stoichiometric) informations around the pre-calculated position. Useful when the automated placement fails to give a clear view of the kinetics / stoichiometry.

Pointer in a selected molecule Arrow Key [move-selection]
Move the currently selected molecules using the keyboard arrow keys.
Pointer in a kinetics/stoichiometry reaction field Arrow Key [move-label]
Move manually the kinetic (or stoichiometric) informations around the pre-calculated position using the keyboard arrow keys. Goes eight times faster when used with the Shift modifier.
Pointer in a kinetics/stoichiometry reaction field Escape [delete-field]
Restore the default (automatic) position of the current kinetics (or stoichiometry) field (the only one enclosed in a dark red rectangle).
Pointer in a kinetics/stoichiometry reaction field Delete [delete-field]
Remove the current kinetics (or stoichiometry) field (the only one enclosed in a dark red rectangle).
Pointer in a text field Delete [delete-char]
Delete the current character (the one in the red box).
Pointer in a text field Left Arrow Key [previous-char]
Move the text cursor to the preceding character.
Pointer in a text field Right Arrow Key [next-char]
Move the text cursor to the next character of the line.
Pointer in a text field Shift+Down Arrow Key [enter-subscript]
Move the text cursor to the subscript position of the current character (creates the subscript if there is none).
Pointer in a text field Shift+Up Arrow Key [enter-superscript]
Move the text cursor to the superscript position of the current character (creates the superscript if there is none).
Pointer in a text field Shift+Left Arrow Key [leave-sub/superscript]
Move the text cursor to the base character of the current subscript / superscript.
Pointer in a text field Ctrl+A [beginning-of-line]
Move the text cursor to the beginning of the line.
Pointer in a text field Ctrl+B [previous-char]
Move the text cursor to the preceding character.
Pointer in a text field Ctrl+D [delete-char]
Delete the current character (the one in the red box).
Pointer in a text field Ctrl+E [end-of-line]
Move the text cursor to the end of the line.
Pointer in a text field Ctrl+F [next-char]
Move the text cursor to the next character of the line.